![PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/88cd5fdbfb2dba16a6b23987d30ad9437ff0c805/7-Table1-1.png)
PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar
Biology | Free Full-Text | Review on the Computational Genome Annotation of Sequences Obtained by Next-Generation Sequencing
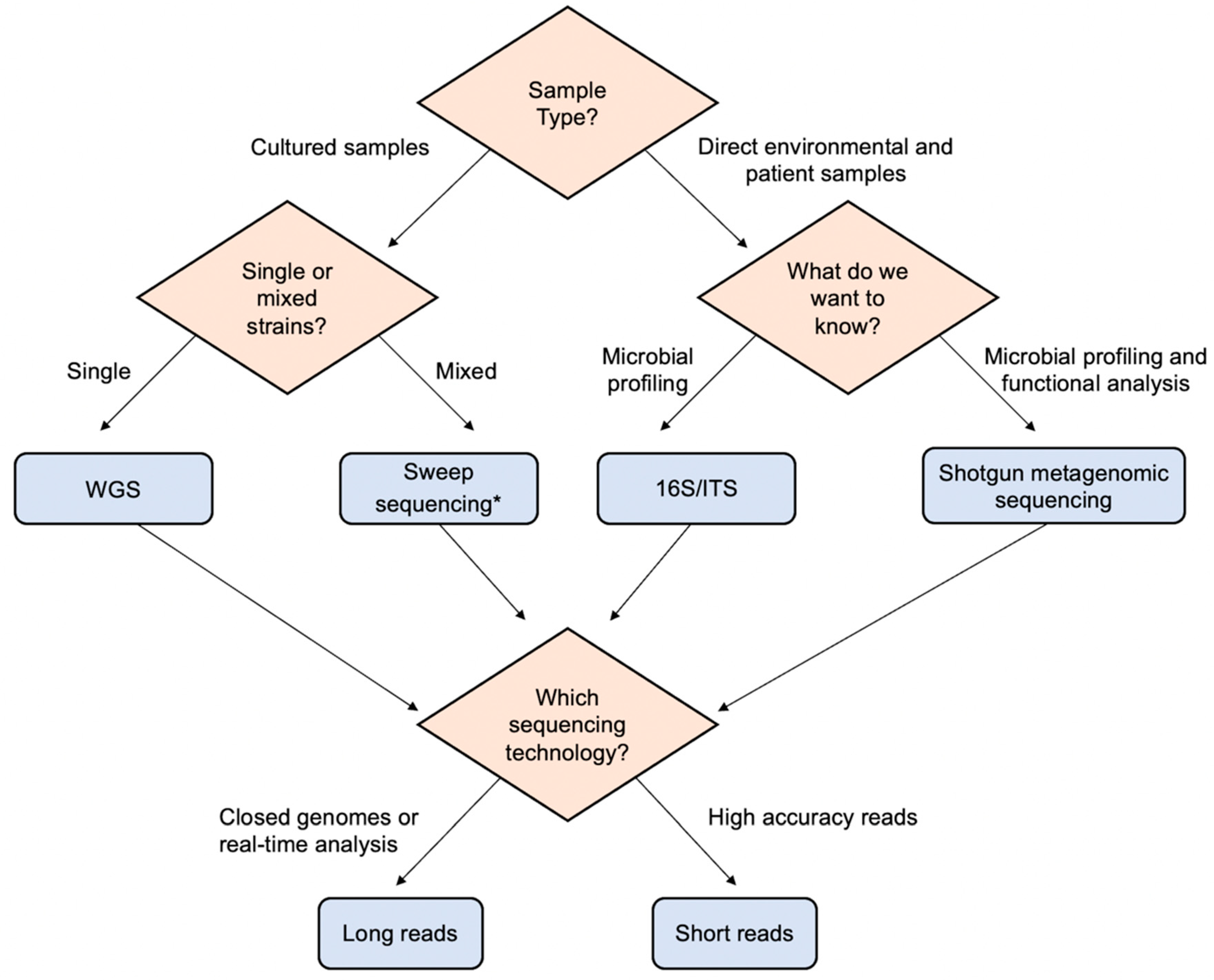
IJMS | Free Full-Text | Combination of Whole Genome Sequencing and Metagenomics for Microbiological Diagnostics

MetaRef comprises automated processing of available microbial genomic... | Download Scientific Diagram
![PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/88cd5fdbfb2dba16a6b23987d30ad9437ff0c805/13-Figure2-1.png)
PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar
![PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/88cd5fdbfb2dba16a6b23987d30ad9437ff0c805/12-Figure1-1.png)
PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar
Metagenomics workflow for hybrid assembly, differential coverage binning, metatranscriptomics and pathway analysis (MUFFIN) | PLOS Computational Biology

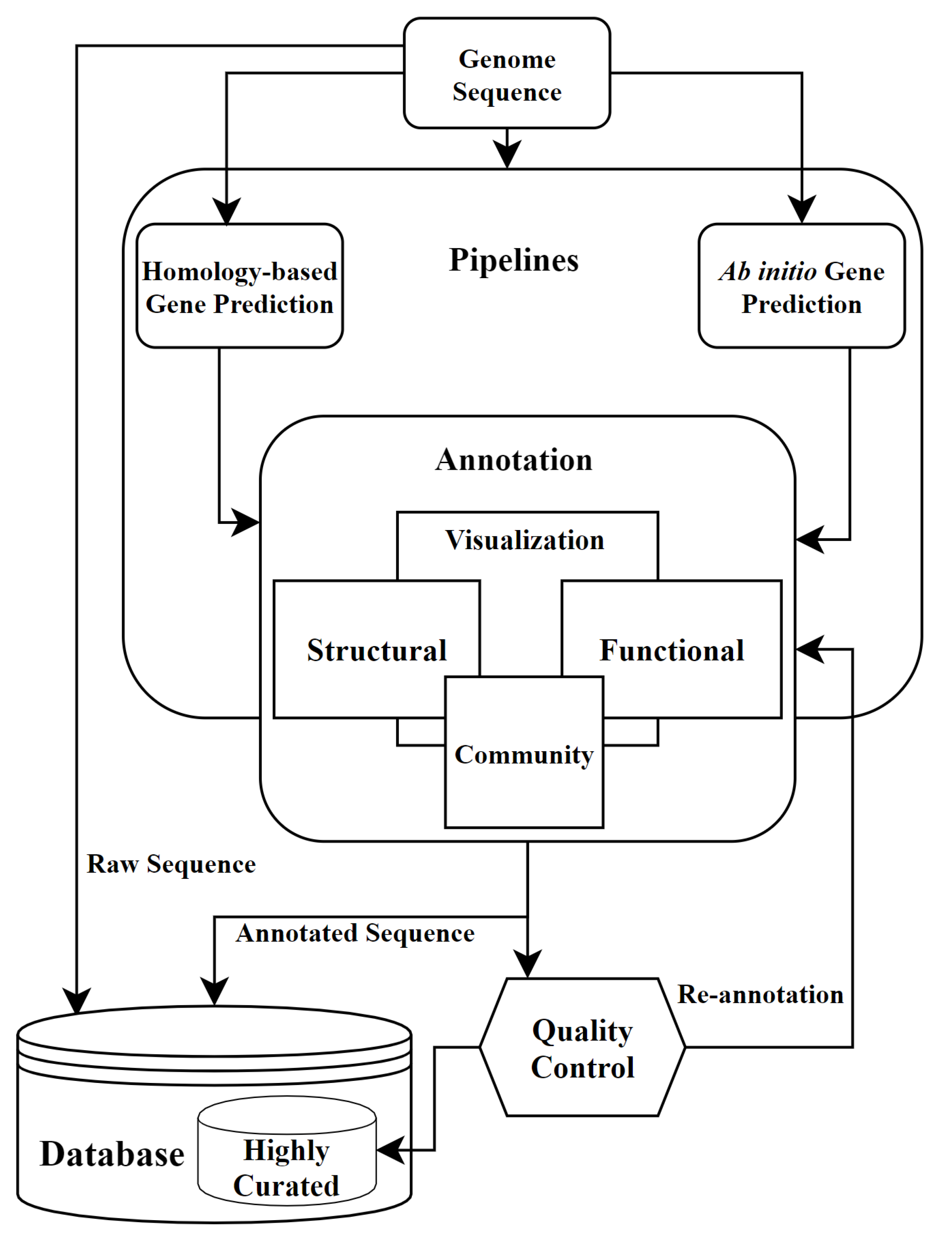
Genome (re‐)annotation and open‐source annotation pipelines - Siezen - 2010 - Microbial Biotechnology - Wiley Online Library
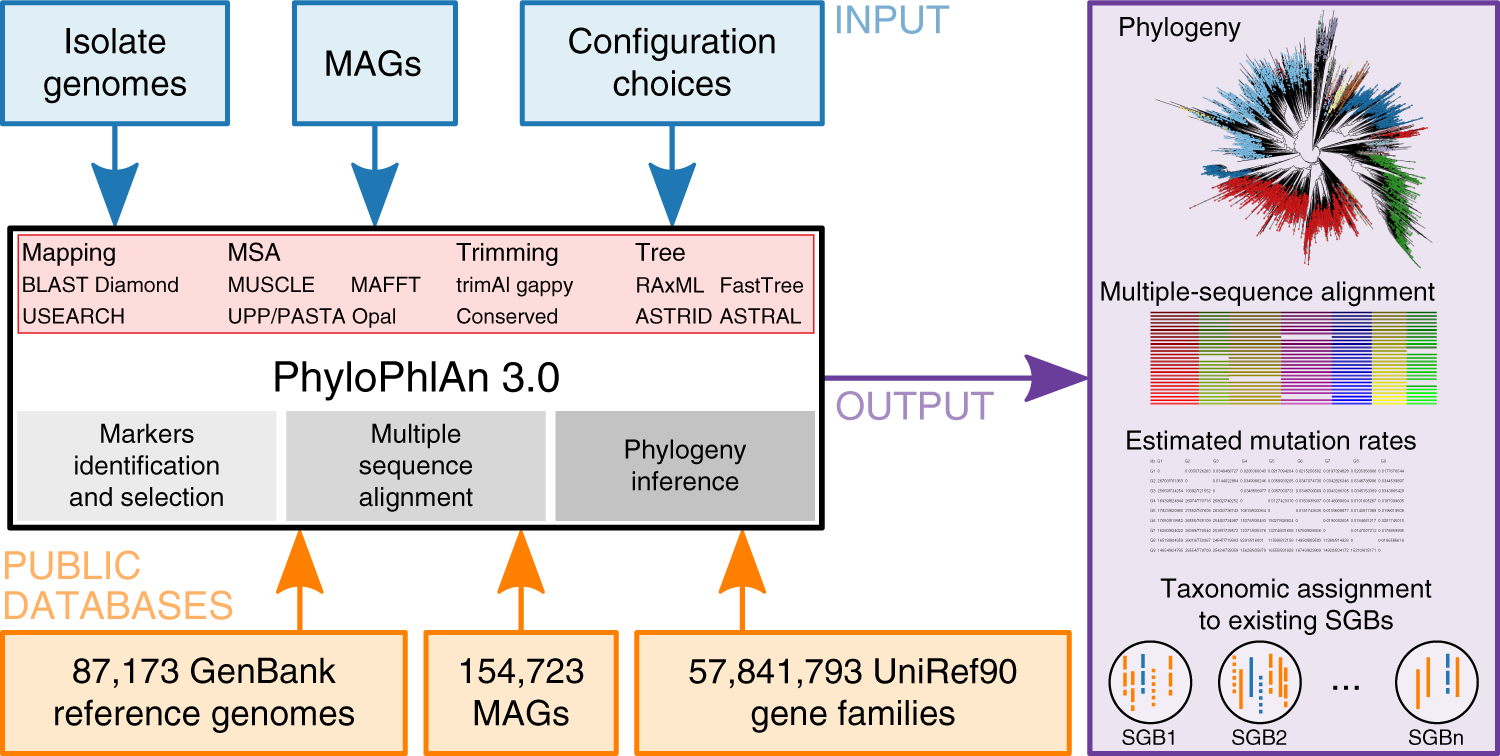
Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0 | Nature Communications
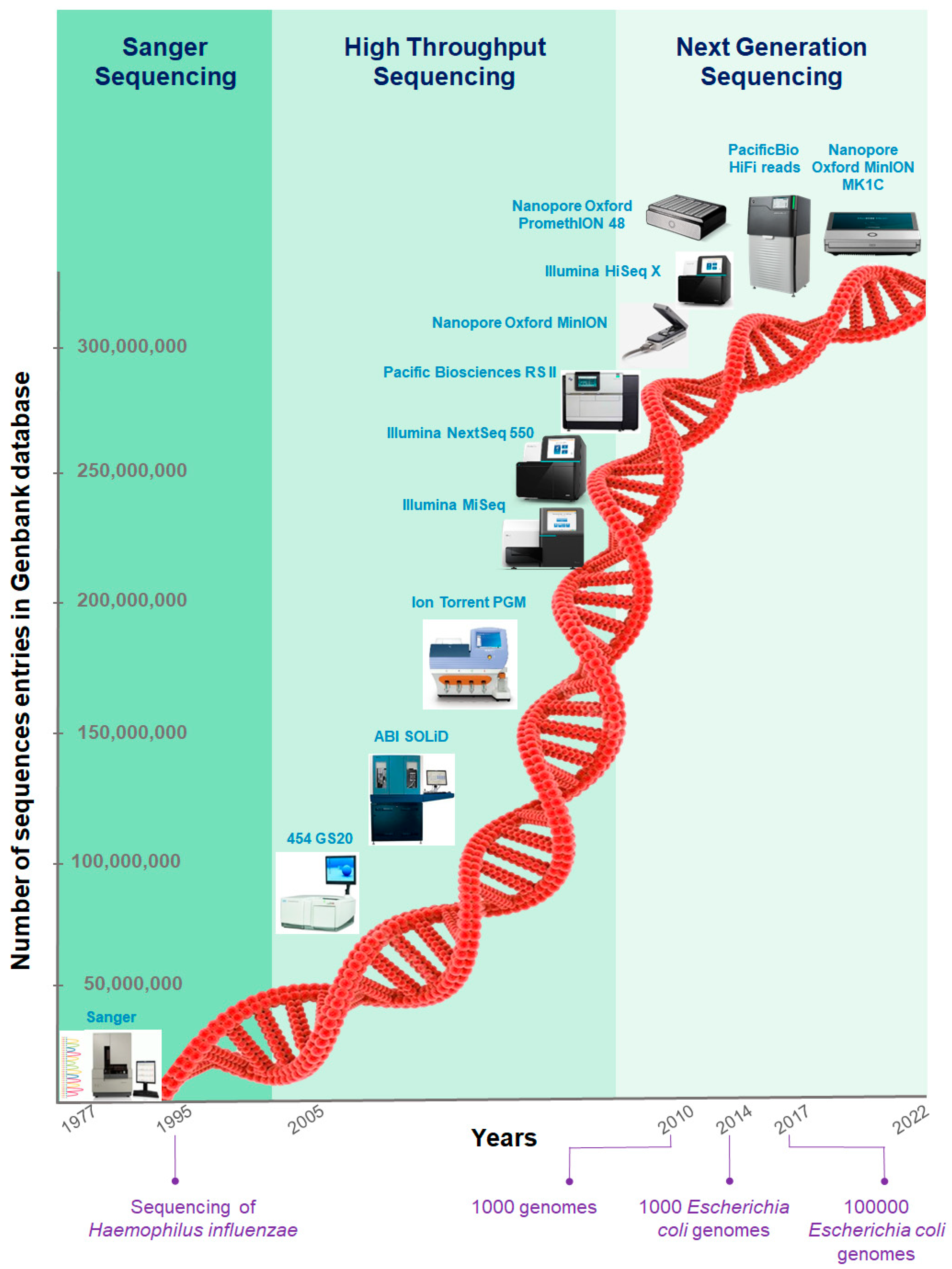
IJMS | Free Full-Text | Application and Challenge of 3rd Generation Sequencing for Clinical Bacterial Studies

Ten Years of Maintaining and Expanding a Microbial Genome and Metagenome Analysis System: Trends in Microbiology

Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen | Nature Communications
![PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/88cd5fdbfb2dba16a6b23987d30ad9437ff0c805/9-Table2-1.png)
PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar
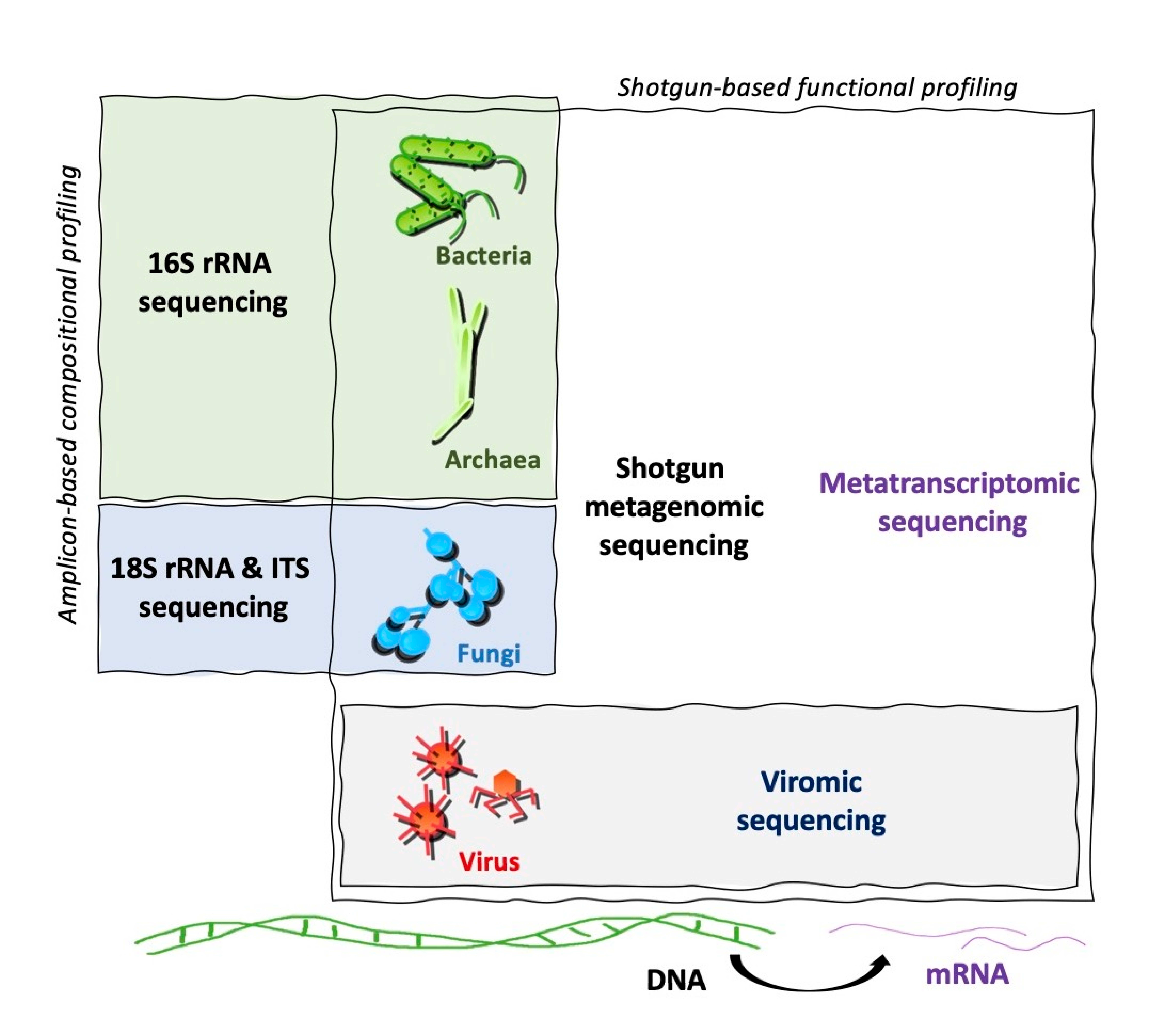
Biomolecules | Free Full-Text | An Introduction to Next Generation Sequencing Bioinformatic Analysis in Gut Microbiome Studies

Genome-Resolved Metagenomics Is Essential for Unlocking the Microbial Black Box of the Soil: Trends in Microbiology
![PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/88cd5fdbfb2dba16a6b23987d30ad9437ff0c805/14-Figure3-1.png)
PDF] Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes | Semantic Scholar

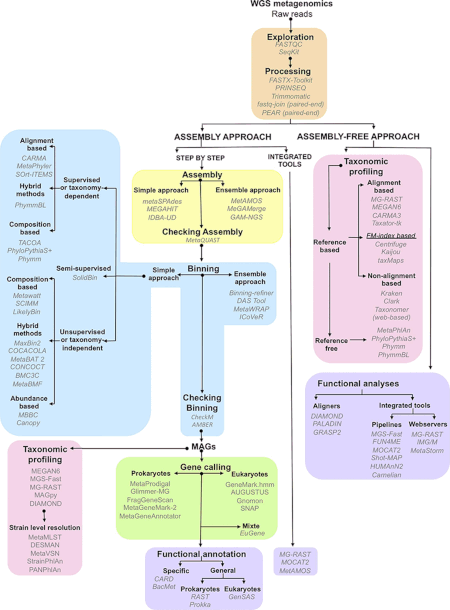
A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data - ScienceDirect
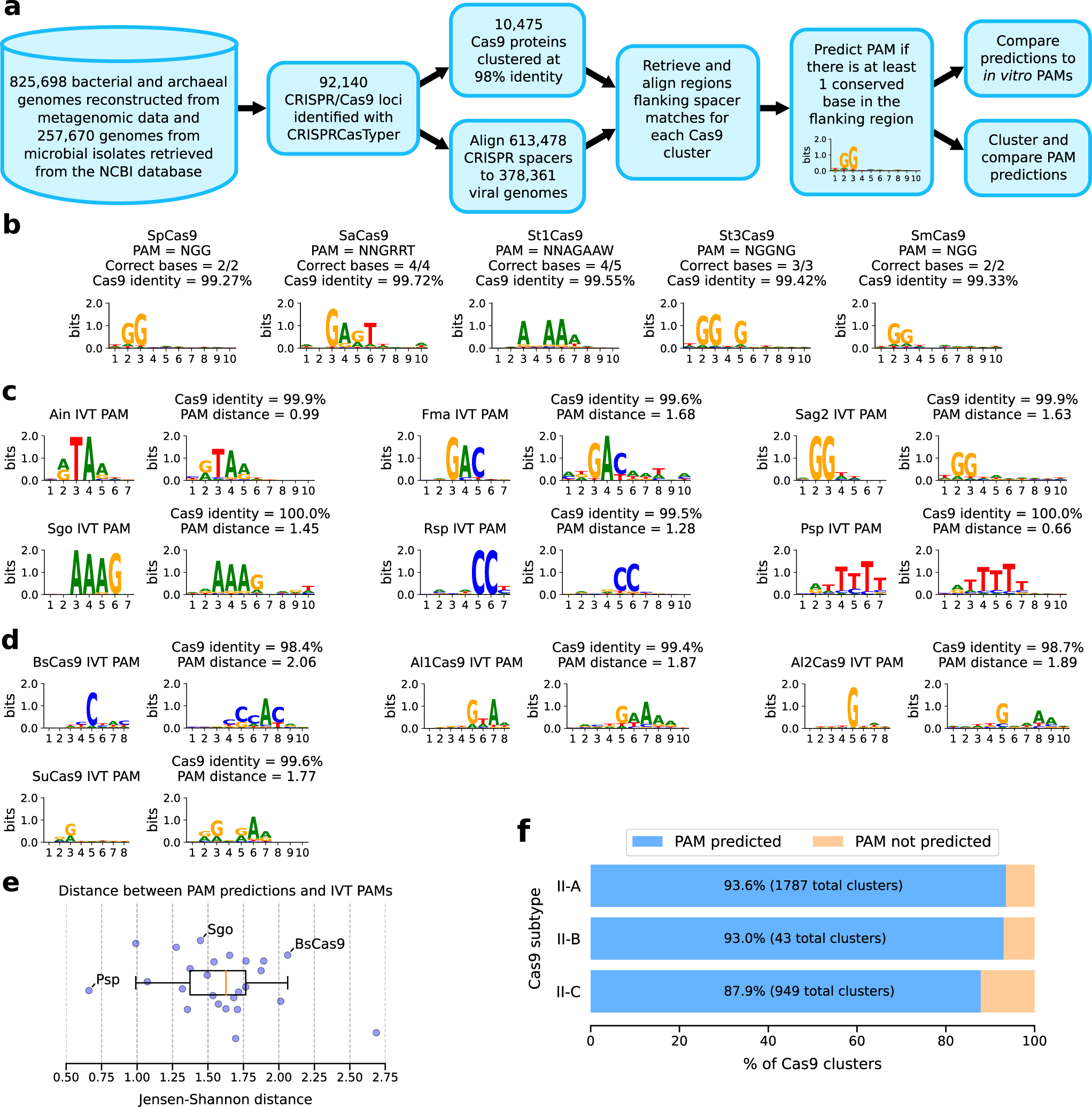
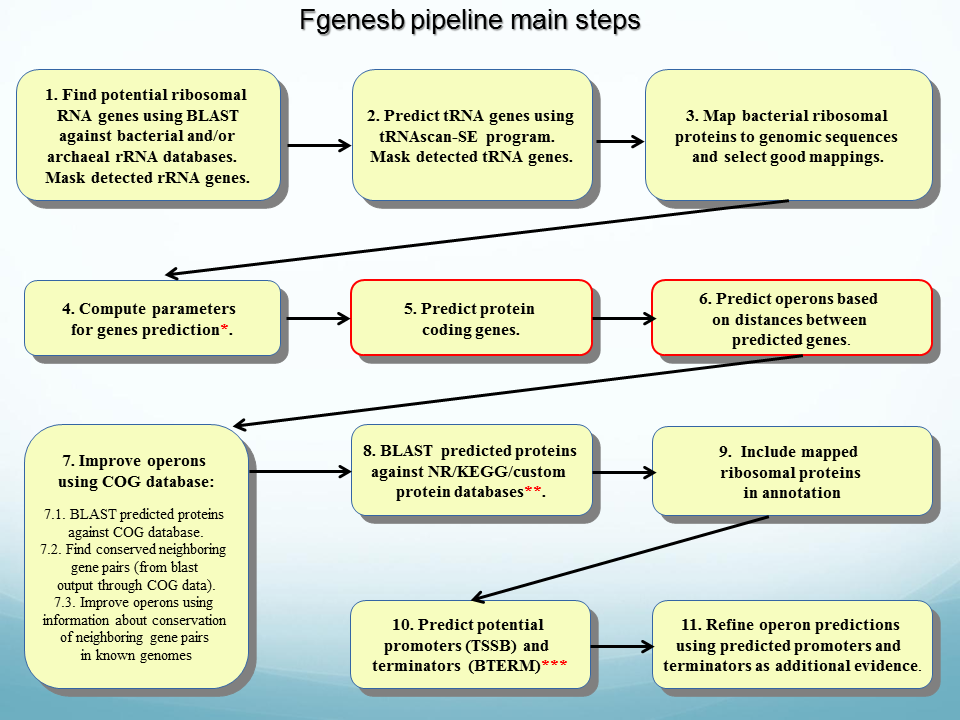


![DATMA: Distributed AuTomatic Metagenomic Assembly and annotation framework [PeerJ] DATMA: Distributed AuTomatic Metagenomic Assembly and annotation framework [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/9762/1/fig-3-full.png)
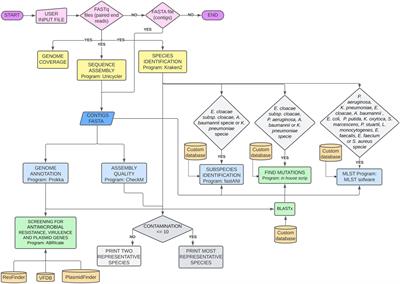
![DATMA: Distributed AuTomatic Metagenomic Assembly and annotation framework [PeerJ] DATMA: Distributed AuTomatic Metagenomic Assembly and annotation framework [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/9762/1/fig-1-full.png)